Annotation of Psyllid Genome
We are annotating the genome of Diaphorina citri using the Apollo system that is hosted at Boyce Thompson Institute. Tom D'Elia from Indian River State College and Mirella Flores and Surya Saha from the Mueller lab at Boyce Thompson Institute are leading this initiative. Teresa Shippy from Kansas State University Bioinformatics Center and Joshua Benoit are the consulting expert annotators.
This project includes annotators across multiple sites including Kansas State University, Indian River State College, University Of Cincinnati and Cornell University. We are using a Basecamp project to organize our activities. We have training webinars and regular online lab meetings. You are welcome to join us. Please let us know if you are interested using the contact form. Please include information about your annotation experience and genes of interest, if any. We will add you to the Basecamp project and invite you to the next annotation video conference.
This project includes annotators across multiple sites including Kansas State University, Indian River State College, University Of Cincinnati and Cornell University. We are using a Basecamp project to organize our activities. We have training webinars and regular online lab meetings. You are welcome to join us. Please let us know if you are interested using the contact form. Please include information about your annotation experience and genes of interest, if any. We will add you to the Basecamp project and invite you to the next annotation video conference.
A screenshot of the WebApollo genome annotation platform
State of Psyllid Annotation
Student publications in GigaByte
These papers are a part of a series of Data Releases crediting the outputs of a student-focused and community-driven manual annotation project for curating gene models and, if required, correcting anomalies in the genome assembly. With the aim of generating a high quality annotation for a better understanding of the biology of this important vector and pathogen-induced changes in gene regulation to aid the development of new strategies in fighting the disease. A total of eleven papers have been published as a part of this series. These will be included in the final Official Gene Set v3.0.
Official Gene Set beta v3.0 released (December 10 2019)
Diaci v3.0 is the current genome version and is being used for annotation on Apollo. Please get in touch using the contact form for write access. The Official Gene Set v3.0 with 19,049 genes is based on Pacbio Isoseq and Illumina RNAseq evidence. All public RNAseq data has been mapped to this genome. We have also mapped Pacbio Isoseq and Illumina RNAseq reads to assist in identification of correct splice sites. Gene models from OGS v1.0, OGS v2.0, De novo transcriptome, MCOT v1, NCBI Gnomon (100 and 101) and other insect genomes have also been made available as evidence tracks. You can download the data from our FTP site. We also provide functional annotation of the proteins in Official Gene Set beta version 3 performed using Rapid Functional Annotation pipeline. Slides describing the results of the functional annotation are available online.
Official Gene Set v2.0 released (March 2018)
Diaci v2.0 is the current genome version and is being used for annotation on Apollo. Please get in touch using the contact form for write access. The Official Gene Set v2.0 with 20,793 genes is based on Pacbio Isoseq and Illumina RNAseq evidence. All public RNAseq data has been mapped to this genome. We have also mapped Pacbio Isoseq and Illumina RNAseq reads to assist in identification of correct splice sites. Gene models from OGS v1.0, MCOT v1, NCBI Gnomon (100 and 101) and other insect genomes have also been made available as evidence tracks. You can download the data from our FTP site.
Diaci1.1 is the previous official version of the Diaphorina citri genome. The contig N50 for this assembly is 38 kb. It was submitted to NCBI and called NCBI-diaci1.1. You can access it here. It was annotated with the NCBI Gnomon pipeline.
The Maker annotations from the Diaci1.0 version of the genome have also been mapped to the current version. The mapping program looked for exact matches between the two assemblies, accounting for stretches of the assembly that were converted to N's. It created an 'alignment file', which was used to perform the re-mapping of features in gff3 format. Maker annotations that mapped to sequence that did not exist in Diaci1.1 or only contained N's in the NCBI-approved assembly were discarded. The Maker annotations that were on retained sequence were preserved in their original format, except for their location (scaffold name, coordinates, and strand if appropriate). 37 Maker gene models from Diaci1.0 did not map to Diaci1.1. Both the Maker annotations and NCBI annotations are available as evidence in Web Apollo.
Official Gene Set v1.0 published (March 2017)
The manual annotation paper describes the workflow for the curation of genes and specific gene families in developmental, physiological, RNAi regulatory, and immunity-related pathways in this gene set. This community effort produced 530 manually curated gene models across developmental, physiological, RNAi regulatory, and immunity-related pathways. As previously shown in the pea aphid, RNAi machinery genes putatively involved in the microRNA pathway have been specifically duplicated. The comprehensive MCOT transcriptome enabled us to identify a number of gene families that are either missing or misassembled in the draft genome. In order to develop biocuration as a training experience, we included undergraduate and graduate students from multiple institutions, as well as experienced annotators from the insect genomics research community. The resulting gene set (OGS v1.0) combines both automatically predicted and manually curated gene models. Please see the supplementary notes and annotation updates for individual gene reports and protein sequences.
Training
There are many training resources available. Monica Munoz-Torres has put together a comprehensive guide on the manual curation process. A recent version of her training slides can be found here. Monica Poelchau at NAL created a tutorial for the new BLAST interface.
Annotation Links
- WebApollo
- The genome page for Diaphorina citri is available here.
- The organism page for Diaphorina citri at i5k is available here.
- New annotators can register to annotate here.
- The data files for annotation are available on the FTP site.
We are using annotation rules adapted from the i5k pilot. The annotators will use the replaced models field to state what model they are replacing with each annotation. Monica Poelchau at the NAL created the following guide to explain the rationale and details of this process.
The Diaphorina citri data that National Agricultural Library hosts can be downloaded from here. The Maker annotations re-mapped to the NCBI-diaci1.1 assembly are available here.




Annotation Updates
C-Type Lectins
Report and protein sequences
C-Type Lectins(CTLs) are calcium dependent extracellular proteins involved in recognition of glycans. A total of 10 models were found and annotated. Refer to C-type lectin gene report for further details. The Basecamp page and FTP site contain the data sets and instructions for annotation.
Relish-like Proteins
Report and protein sequences
Relish-like proteins are transcription factors and are inlvolved in Imd pathway. The Basecamp page and FTP site contain the data sets and instructions for annotation.
Galactoside-Binding Lectins
Report and protein sequences
Galactoside-Binding Lectins (GALEs) or galectins bind to beta-galactoside sugars. GALEs are implicated in innate immunity as they might be involved in microbial recognition and/or phagocytosis. The Basecamp page and FTP site contain the data sets and instructions for annotation.
Fibrinogen-Related Proteins annotation
Report and protein sequences
Fibrinogen-Related Proteins (FREPs) are implicated in bacteria binding, enhancement of antimicrobial activity and interaction with parasite. The Basecamp page and FTP site contain the data sets and instructions for annotation.
TEPs annotation
Report and protein sequences
Thio-Ester containing proteins (TEPs) are involved in pathogen recognition and activation of immune responses. The Basecamp page and FTP site contain the data sets and instructions for annotation.
PGRP annotation
Report
Peptidoglycan Recognition proteins are pattern recognition molecules conserved from insects to mammals and recognize bacteria and their unique cell wall component, peptidoglycan (PGN). The Basecamp page and FTP site contain the data sets and instructions.
CLIP-Domain Serine Proteases
CLIP-Domain Serine Proteases (CLIPs) are large protein family unique to arthropods. CLIP proteases are activated after infection and function in extracellular pathways for proteolytic activation of downstream proteins. The Basecamp page and FTP site contain the data sets and instructions for annotation.
Inhibitors of Apoptosis
Inhibitors of Apoptosis (IAP) protein family binds to caspases which are involved in apoptotic cell death program. IAPs are also been shown to be down-regulated in cancer cells leading to tumor formation. The Basecamp page and FTP site contain the data sets and instructions for annotation.
Catalases genes annotation
Catalases (CATs) are involved in the conversion of hydrogen peroxide to water and oxygen. The Basecamp page and FTP site contain the data sets and instructions.
Autophagy genes annotation
Autophagy genes (Atg) are involved in the formation of autophagosome leading to degradation of internalized pathogens. The Basecamp page and FTP site contain the data sets and instructions.
Prophenoloxidases
Prophenoloxidase are involved in innade defense system which play major role in melanization. The Basecamp page and FTP site contain the data sets and instructions for annotation.
Peroxidases
Peroxidases produce local reactive oxygen species as a immune response. The Basecamp page and FTP site contain the data sets and instructions for annotation.
MD2-Like Receptors
MD2-Like Receptors (MLs) are extracellular proteins involved in signal transduction pathways by lipid recognition. The Basecamp page and FTP site contain the data sets and instructions for annotation.
CASPAs annotation
Caspase Activators include ARK (Apaf-1 Related Killer) and IAP (Inhibitor of Apoptosis) antagonists. The Basecamp page and FTP site contain the data sets and instructions.
SRRP annotation
Small Regulatory RNA pathway members are important in many cellular processes as well as in immune defence. The Basecamp page and FTP site contain the data sets and instructions.
CASPs annotation
Caspases are proteolytic enzymes that cleave aspartic acid of target proteins. The Basecamp page and FTP site contain the data sets and instructions.
AMP annotation
Genes producing anti-microbial peptides are a critical part of the immune system of the psyllid. The Basecamp page and FTP site contain the data sets for annotating AMPs.
This track contains tblastn matches to the genome for MCOT proteins. MCOT combines gene predictions from Maker, Cufflinks, Oasis and Trinity. It also includes the uniprot annotation if available.
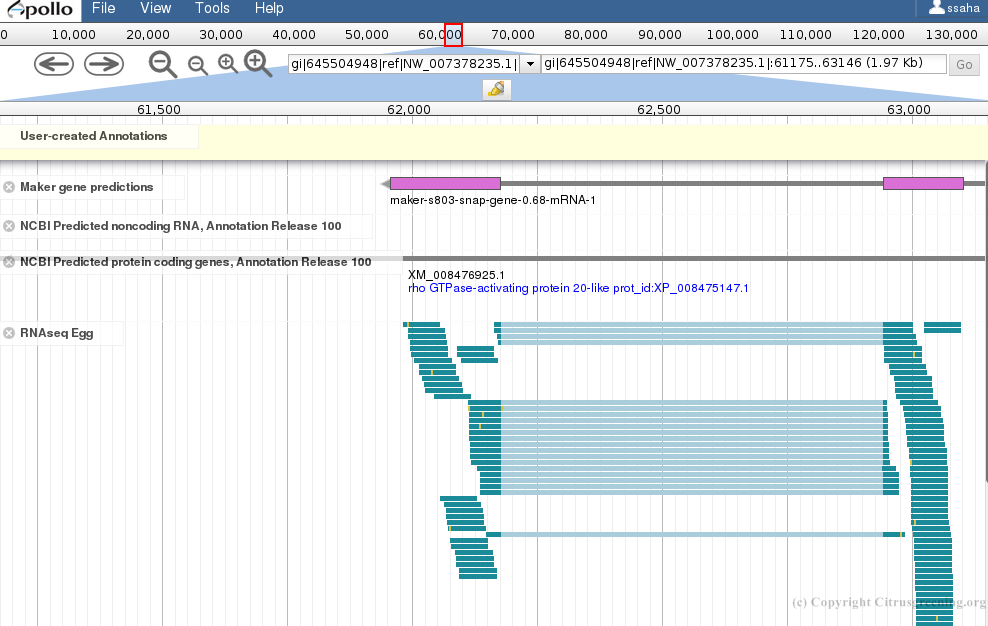
Evidence tracks for RNAseq reads from egg, nymph and adult tissue were added to WebApollo to assist in annotation.
Basecamp site is now available
The project basecamp site is now available. We will use this to coordinate all research activities for the project.